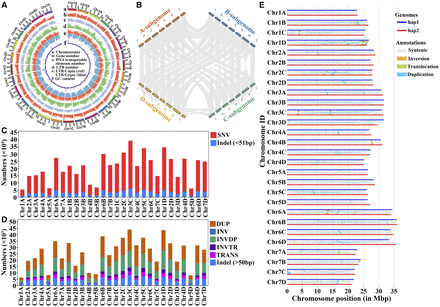
Genome assembly of “Beni hoppe” and variations between hap1 and hap2. (A) Genomic features of the “Beni hoppe” genome FaBen. All statistics were computed for windows of 100 kb. (B) Intergenomic syntenic analysis between A–D subgenomes of FaBen. (C) Number of SNVs and indels (<51 bp) in the same chromosome pair between hap1 and hap2. (D) Number of structural variations in the same chromosome pair between hap1 and hap2. (DUP) duplicated region, (INV) inverted region, (INVDP) inverted duplicated region, (INVTR) inverted translocated region, (TRANS) translocated region. (E) Structural variations in the same chromosome pair between hap1 and hap2.











