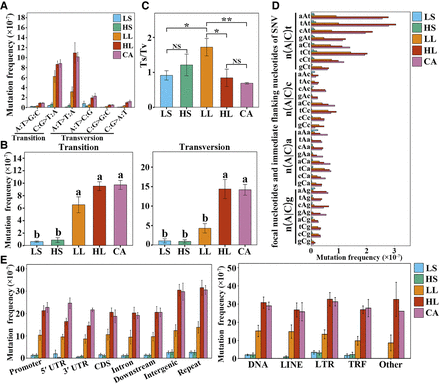
Characterization of the induced SNVs in regenerated woodland strawberry plants. (A) Mutation frequencies (number of mutations per site) of different transition or transversion types in regenerated plants. (B) Mutation frequencies (number of mutations per site) of all transitions and transversions in regenerated plants. Different letters indicate significant differences at P < 0.05 using Tukey's test. (C) Transition/transversion (Ts/Tv) ratios in regenerated plants. (*) P < 0.05, (**) P < 0.01, (NS) not significant, Student's t-test. (D) Neighbor-dependent mutation frequencies (number of mutations per site) at AT and GC bases in regenerated plants. The trinucleotide context-dependent mutation frequency is shown for each treatment. Uppercase indicates mutation site and lowercase indicates immediate flanking nucleotides and n indicates a, t, c, or g. (E) Mutation frequencies (number of mutations/total length of each region type) of SNVs in different genomic regions or transposon families. (Promoter) 2 kb upstream of transcription start site (TSS), (UTR) untranslated region, (Downstream) 2 kb downstream from transcription termination site (TTS), (Repeat) transposable element or tandem repeat sequence, (TRF) tandem repeat sequence, (Other) unclassified transposon. Data are the mean ± SEM.











