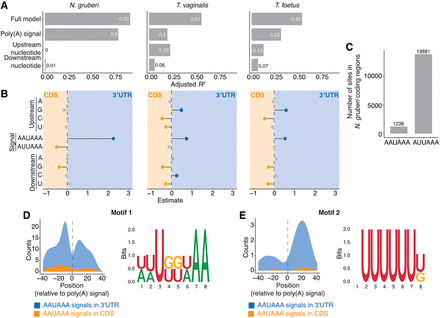
Flanking nucleotides play a role in poly(A) signal recognition in other Metamonada, but not N. gruberi. (A) Variance explained by the linear model applied to AWUAAA-containing k-mers scores from gkmSVM classifier. In the outgroup organism N. gruberi, signal sequence is the main determinant, whereas in T. vaginalis and T. foetus from Metamonada upstream nucleotides have substantial contributions. (B) Beta-coefficient values from the linear model form A. (C) AAUAAA and AUUAAA hexamer occurrence in the coding sequences of N. gruberi. Occurrence was calculated independent of the reading frame. (D,E) Auxiliary elements help specify genuine AAUAAA poly(A) signals in N. gruberi, putative upstream (D) and downstream (E) motifs and their quantifications around AAUAAA poly(A) signals.











