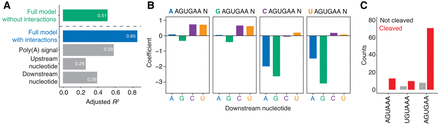
Functional poly(A) signals in the majority of WGURAA hexamers in coding sequences in G. muris. (A) Variance explained by the linear model applied to WGURAA-containing k-mers scores from gkmSVM classifier by the full model, upstream nucleotide, poly(A) signal, and downstream nucleotide in G. muris. Explained variance was measured as an adjusted R2 value. The green bar represents a full linear model without interactions, blue—with interactions, gray—components of the model with interactions. Beta-coefficient values are found in Supplemental Figure S4A. (B) Influence of upstream and downstream nucleotide interaction to AGUGAA hexamers classification. Upstream nucleotide is color-coded at the top of each barplot. Data for AGUAAA and UGUAAA hexamers are shown in Supplemental Figure S4B and C, respectively. (C) Quantification of premature cleavage events in G. muris coding sequences by hexamer identity. Among 199 WGURAA hexamers in G. muris coding sequences, 106 had sufficient read support and 91 were cleaved.











