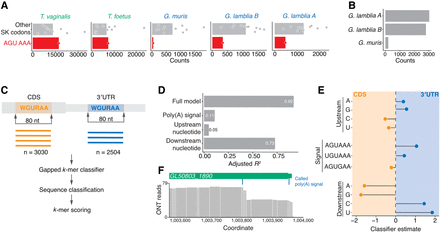
Importance of 3′ adjacent nucleotide of the WGURAA hexamer for distinguishing poly(A) signals from open reading frame hexamers. (A) Occurrence of AGU AAA dicodon in comparison to other Ser-Lys encoding dicodons in coding sequences within Metamonada clade. Dicodon occurrence was calculated only in frame. Each point represents one dicodon combination and the bar shows the mean count value for all compared combinations. AGU AAA was shown to be depleted in the Giardia genus, but not in other species. Data for AAA AGU, UGU AAA, and AGU GAA dicodons are shown in Supplemental Figure S3A. (B) WGURAA hexamer occurrence in the coding sequences of Giardia species. Occurrence was calculated independent of the reading frame. (C) Schematic representation of the machine learning approach to distinguish WGURAA sequences in 3′ UTRs versus coding sequences. WGURAA sequences were extracted from coding sequences and 3′ UTRs and together with 37 flanking nucleotides from both sides (80-mer) put into a gapped k-mer support vector machine classifier, which performed sequence classification and k-mers scoring. (D) Variance explained by the linear model applied to WGURAA-containing k-mers scores from gkmSVM classifier by the full model, upstream nucleotide, poly(A) signal, and downstream nucleotide in G. lamblia. Explained variance was measured as an adjusted R2 value. Data for G. lamblia B are shown in Supplemental Figure S3C. (E) Beta-coefficient values from the linear model applied to WGURAA-containing k-mers scores from gkmSVM classifier, corresponding to the upstream nucleotide, poly(A) signal, and downstream nucleotide in G. lamblia. Data for G. lamblia B are shown in Supplemental Figure S3B. (F) Example of G. lamblia A gene GL50803_1890 where a hexamer in the coding sequence was misclassified by gkmSVM. Premature cleavage after the hexamer inside the coding sequences indicated as coverage drop was observed. Similar example from G. lamblia B is shown in Supplemental Figure S3D.











