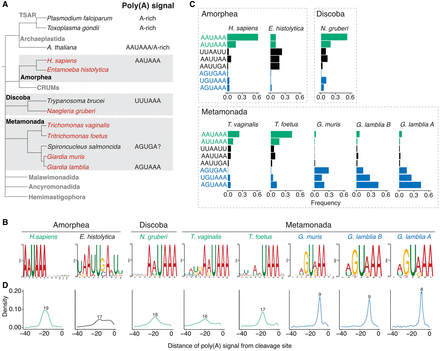
Characterization of poly(A) signals in diverse protists. (A) Simplified view of phylogenetic tree indicating evolutionary relationships between sequenced protist species (highlighted red) and other organisms. Known poly(A) signal sequences are shown next to the organism's name. (B) MEME analysis of sequence motifs enriched in the regions 40 nt upstream of the cleavage sites. Analysis was performed for all sequenced protist species and human annotated 3′-UTR sequences. Sequences representing previously described unusual AGURAA G. lamblia poly(A) signals were enriched within the Giardia genus. Sequences with enriched AWUAAA-like motifs are highlighted in green, AGURAA-like sequences in blue, and others in black. (C) Quantification of hexamers frequencies in annotated 3′ UTRs in the regions 40 nt upstream of the cleavage sites, based on the enrichment results from B. Color schemes the same as in B. (D) Distribution of the distances between poly(A) signals and cleavage sites. Poly(A) signals in the Giardia genus are positioned closer to the cleavage sites than in other organisms. Most common distance is shown above the plot. Color scheme is the same as in B.











