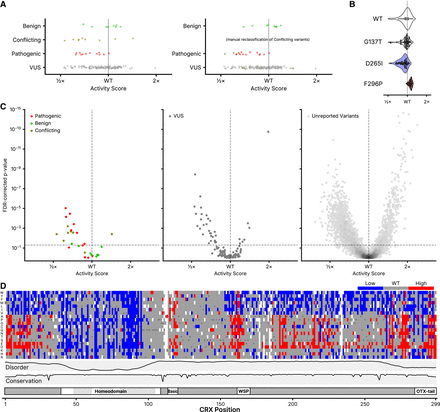
Classification of CRX variants. (A) DMS activity scores for variants reported to ClinVar in each of the indicated classes (Pathogenic includes “Pathogenic” and/or “Likely pathogenic”; Benign includes “Benign” and/or “Likely Benign”). On the right, “Conflicting” variants are shown reclassified based on the modal reported ClinVar classification. (B) Barcode-level activity measurements for wild-type CRX and the indicated representative wild-type-like (p.G137T), low-activity (p.D265I), and high-activity (p.F296P) variants. Each black dot represents a unique barcoded construct; not shown for wild-type CRX due to it being barcoded thousands of times. (C) Volcano plots showing classifications for the indicated ClinVar variants (left and middle panels) or all other variants not yet reported in ClinVar (right panel). FDR-corrected P-values were computed from a two-sample K–S test comparing each variant's barcode-level measurements to those of wild-type CRX, as visualized in (B). The horizontal line corresponds to an FDR-corrected P-value of 0.05; the vertical line corresponds to a normalized activity score of 1 (wild-type). For visualization purposes, the y-axis is clipped to 10−15; 13 variants are hidden with activity scores greater than wild-type and P-values up to 10−26. (D) Quantized DMS activity scores for each variant, coloring low- and high-activity variants using the significance cutoffs shown as dotted lines in (C). Disorder, domains, and conservation are shown as in Figure 1.











