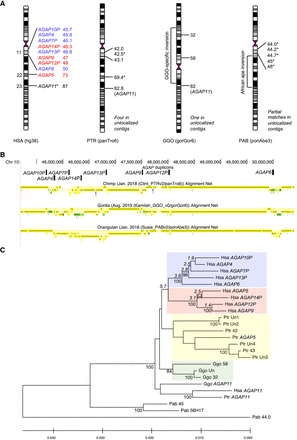
Genomic organization and phylogeny of Chromosome 10 AGAP copies in great apes. (A) Chromosome location (in Mbp) of AGAP copies in human, chimpanzee, gorilla, and orangutan reference genomes according to the most recent releases. In the common ancestor of African apes, a pericentric inversion moved AGAP11 outside the pericentromeric region. Copies with an asterisk (*) are shorter than the others and are thus incomplete. Human AGAP copies are colored according to the clade in the phylogenetic tree shown in C. (B) Pairwise genomic sequence alignments of the q-arm pericentromeric region of human Chromosome 10 with great ape genomes (UCSC track of primate net alignments). The location of AGAP duplicons is shown. (C) Phylogeny of Chromosome 10 AGAP copies in great apes. The tree was inferred by using the Maximum Likelihood method and Kimura 2-parameter model. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. All positions containing gaps and missing data were eliminated (complete deletion option). There were a total of 4961 positions in the final data set. Bootstrap values >75 are shown next to significant nodes. Timing estimates in millions of years of split events between human AGAP copies are shown in italics.











