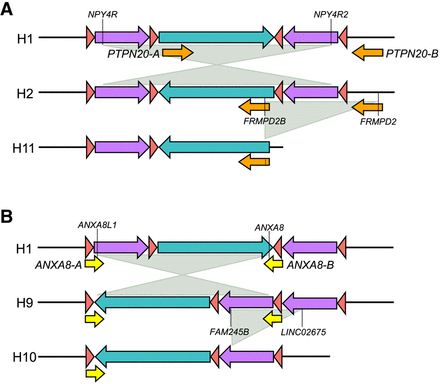
Inversion haplotypes generate new predispositions to copy-number variations. (A) Schematic of H2 and H11 haplotypes. The H2 haplotype differs from H1 by an inversion mediated by NPY4R inverted duplicons (purple arrows). Following this inversion, the PTPN20 duplicons (orange arrows) become in direct orientation and mediate the deletion identified in H11. Genes mapped at the breakpoints are specified. (B) Schematic of H9 and H10 haplotypes. The H9 haplotype differs from H1 by an inversion mediated by ANXA8 inverted duplicons (yellow arrows). Following this inversion, the NPY4R duplicons (purple arrows) become in tandem configuration and direct orientation and mediate the deletion of one copy in H10 haplotype. Genes mapped at the breakpoints are specified.











