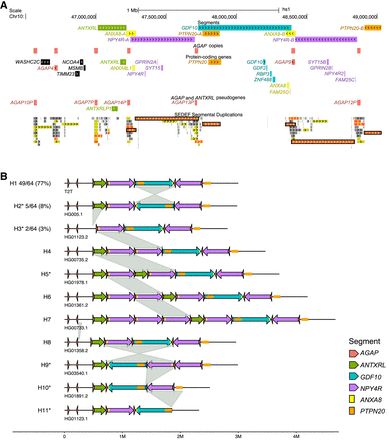
Structural variation at the 10q11.22 chromosomal region. (A) Annotation of segments, genes, and SDs. View in the UCSC Genome Browser on human T2T-CHM13 v2.0 of the region Chr 10: 46,500,000–49,100,000. The following segments are drawn: ANTXRL (green), NPY4R-A/B (violet), ANXA8-A/B (yellow), PTPN20-A/B (orange), and GDF10 (blue). ANXA8-A overlaps NPY4R-A whereas ANXA8-B is just outside the NPY4R-B segment. The location of AGAP copies (protein-coding genes and pseudogenes) is shown with red boxes. Protein-coding genes and the ANTXRL pseudogene are indicated, with the text colored according to the segment to which they belong. At the bottom, SD annotation with colors reflecting the level of similarity: light to dark gray for 90%–98%; yellow for 98%–99%; orange for similarity >99% (Numanagic et al. 2018; Vollger et al. 2022). Black rectangles mark the three pairs of inverted SDs that overlap the NPY4R, ANXA8, and PTPN20 segments, respectively. (B) Schematic of 11 alternative haplotypes identified in 64 haploid genomes. Numbers next to H1, H2, and H3 haplotypes represent their frequency. All other haplotypes were identified in a single genome. H1 corresponds to the T2T-CHM13 haplotype and is the most common. Asterisks denote haplotypes validated by the GAVISUNK tool. Gray links between haplotypes highlight the underlying structural variation.











