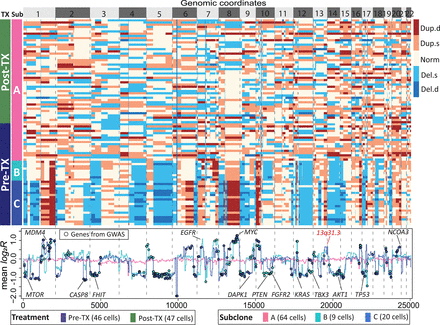
Subclone clustering of the KTN126 patient using FLCNA. Cell clusters and copy number profile with different CNA states (deletion of double copies [Del.d], deletion of a single copy [Del.s], normal/diploid [Norm], duplication of a single copy [Dup.s], and duplication of double copies [Dup.d]) were generated using FLCNA. The mean logarithm transformation of ratio between normalized read counts and its sample-specific mean (log2R) was provided for each cluster. Shared CNAs identified using FLCNA were matched to significant genes from the genome-wide association studies (GWASs) in the NHGRI-EBI GWAS Catalog. (Pre-TX) Pretreatment, (Post-TX) posttreatment.











