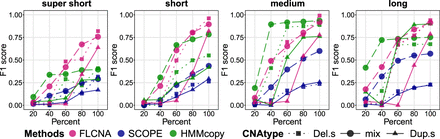
Accuracy of CNA detection in simulated data with five clusters. CNA calls were generated by FLCNA, SCOPE, and HMMcopy, respectively. For each of five clusters, we added 50 CNA segments with varied lengths (supershort: two to five markers; short: five to 10 markers; medium: 10 to 20 markers; and long: 20 to 35 markers) and varied CNA proportions (20%, 40%, 60%, 80%, 100%), respectively, to the background signal. Deletion of a single copy (Del.s), mixed CNA states (mix), and duplication of a single copy (Dup.s) were spiked in separately. F1 score was used to evaluate the performance of CNA detection for each method.











