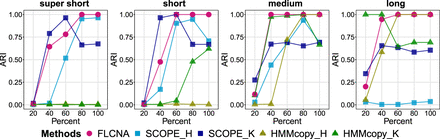
Accuracy of clustering in simulated data with five clusters and mixed CNA states. Clustering results from FLCNA were compared with existing methods (i.e., SCOPE and HMMcopy) coupled with different clustering methods. For each of five clusters, we added 50 CNA segments with varied lengths (supershort: two to five markers; short: five to 10 markers; medium: 10 to 20 markers; and long: 20 to 35 markers) and varied CNA proportions (20%, 40%, 60%, 80%, 100%), respectively, to the background signal. Signals of mixed CNA states (i.e., Del.d, Del.s, Norm, Dup.s, and Dup.d) were spiked in. (ARI) Adjusted Rand index, (SCOPE_H) SCOPE_hierarchical, (SCOPE_K) SCOPE_K-means, (HMMcopy_H) HMMcopy_hierarchical, and (HMMcopy_K) HMMcopy_K-means.











