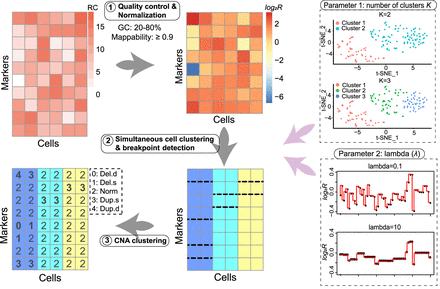
Analysis workflow of FLCNA. FLCNA first implements a quality-control procedure based on GC content and mappability. A two-step median normalization approach sequentially removes the effect of each bias. Data are then scaled by their cell-specific mean and undergo a logarithm transformation (log2R). With log2R, FLCNA clusters subclones and simultaneously detects shared breakpoints using a Gaussian mixture model with a fused lasso penalty term. Finally, based on these shared breakpoints in each cluster, segments for each cell are clustered into five different CNA states: deletion of double copies (Del.d), deletion of a single copy (Del.s), normal/diploid (Norm), duplication of a single copy (Dup.s), and duplication of double copies (Dup.d). There are two hyperparameters in the FLCNA model, the number of cell clusters K and a tuning parameter λ, which controls the number of breakpoints.











