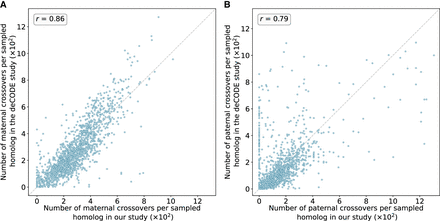
Number of maternal (A) and paternal (B) crossovers per sampled homolog in genomic bins compared between our study and published genetic maps from deCODE. Data from deCODE were obtained from Halldorsson et al. (2019). Crossovers were identified as transitions between regions of “matched” and “unmatched” haplotypes, in which each region included at least 15 genomic windows and a z-score of at least 1.96. Pearson correlation coefficients (r) between the deCODE map and our map across genomic bins are reported for each panel (for chromosome-specific comparison, see Supplemental Figs. S9, S10).











