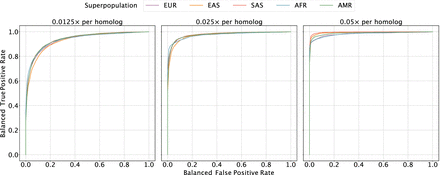
Evaluating the sensitivity and specificity of meiotic crossover detection based on simulation. Using data from the 1000 Genomes Project, we simulated pairs of monosomy 16 and disomy 16, in which half of the pairs possessed matched haplotypes and the other half possessed unmatched haplotypes. Then we divided Chromosome 16 into 45 bins (of ∼2 Mbp) and calculated a balanced ROC curve (see Methods) for each bin, averaging over all the balanced ROC curves to obtain a mean balanced ROC curve. We repeated this procedure over a range of depths of coverage and across sets of samples from all superpopulations of the 1000 Genomes Project, abbreviated as follows: (AMR) admixed American, (AFR) African, (EAS) East Asian, (EUR) European, (SAS) South Asian.











