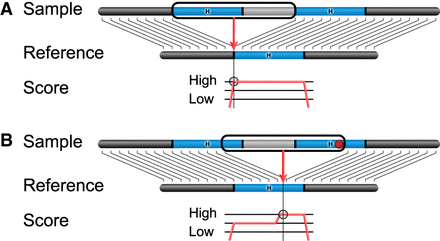
A model of breakpoint differences. SV insertion breakpoints in homologous sequence (“H,” blue) can be placed differently with little effect on the alignment score. (A) In the absence of differences between the homologous loci, aligners typically left-align the breakpoint consistently placing it on the left side of the homologous locus in the reference. (B) When a small polymorphism such as a SNP or indel is in the homologous region (red dot), the alignment score is penalized for the mismatching base. If the breakpoint is shifted so that the polymorphism falls inside the inserted sequence, the penalty is eliminated, and it appears to produce a better alignment. A different representation for this SV insertion is produced in samples with the SNP.











