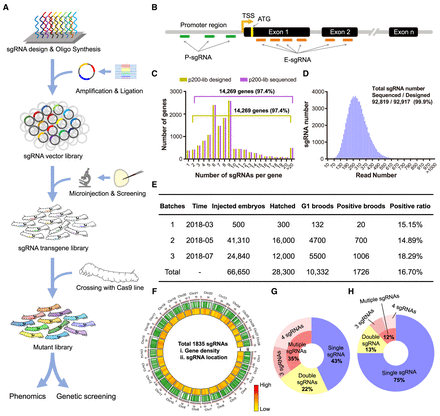
Design and construction of the CRISPR single-guide RNA (sgRNA) library. (A) The strategy of the mutant library construction. (B) The schematic diagram of sgRNA design that targets the promoter region and exon around whole genome of B. mori. The sgRNAs targeting the promoter region were named by P-sgRNA, whereas sgRNAs targeting the exon1 or exon2 were named by E-sgRNA. (C) The sgRNA distribution for the p200-lib of designed and deep sequenced. (D) Distribution of the deep sequenced reads and sgRNA numbers. (E) Summary of microinjection information for B. mori. (F) Distribution of sgRNAs detected by deep sequencing over the B. mori whole genome. (G,H) The number of sgRNAs detected in individuals.











