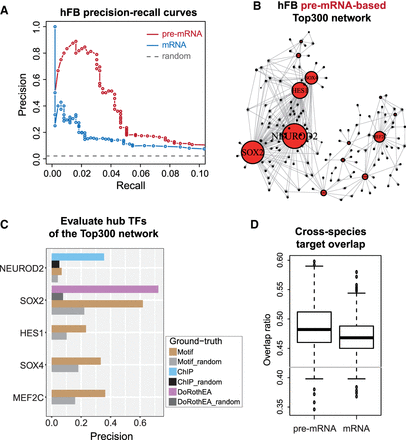
Using the forebrain data sets to demonstrate the improved performance of the pre-mRNA-based method. (A) Precision-recall curves for GRNs inferred by pre-mRNA-based and mRNA-based methods for the human forebrain data set. The ground-truth network is DoRothEA GRN. (B) Inferred GRN for the human forebrain data set using the pre-mRNA-based method. In this network, the edges represent the inferred transcriptional regulation from one TF (transcription factor) to one target gene. The size of the node represents the number of inferred target genes for the TF. 300 interactions (edges) of the highest confidence were shown (i.e., Top300 network). (C) Evaluation of the hub TFs in the pre-mRNA-based Top300 network using different types of ground-truth network. Note that because the DoRothEA ground-truth GRN contains much fewer nodes and edges compared to the Motif ground-truth GRN, only one of the hub TFs could be evaluated using DoRothEA GRN whereas all of them could be evaluated with the Motif GRN. (D) Ground-truth-free comparison between GRNs inferred by pre-mRNA-based and mRNA-based methods using cross-species target overlap. In particular, GRNs from the human forebrain data set (hFB) and the mouse forebrain (mFB) were evaluated, whereby the overlap ratio of top 500 targets in two networks for each TF was calculated. Such overlap ratio represents the cross-species similarity between the two networks, and was compared between pre-mRNA-based and mRNA-based GRNs. N = 372 TFs and P-value = 5 × 10−9 from Wilcoxon test. Gray horizontal line indicates random overlap ratio.











