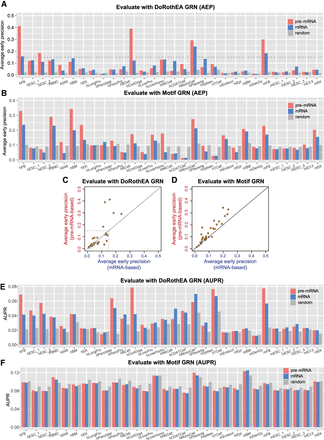
Using experimental single-cell data sets to compare pre-mRNA-based and mRNA-based methods. (A) Comparison between pre-mRNA-based and mRNA-based methods for 30 scRNA-seq data sets of human and mouse cells using DoRothEA GRN as the ground truth. The performance was measured by average early precision (Methods). Precision for a random predictor is shown for comparison. (B) Analogus to A, except that Motif GRN was used as the ground truth. (C) Scatter plots showing the comparison between pre-mRNA-based and mRNA-based methods for 30 scRNA-seq data sets of human and mouse cells using DoRothEA GRN as the ground truth. Diagonal line denotes equal performance. Dashed lines denote precision value at 0.15. (D) Scatter plots analogous to C, except that Motif GRN was used as the ground truth. (E,F) Analogous to A and B, except that AUPR was used as the metric.











