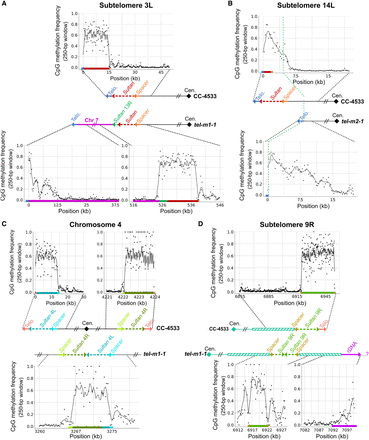
Methylation frequency of new extremities and displaced Sultan elements. (A–D) Analysis of 5mC frequency at CpG sites using reads that unambiguously spanned the indicated regions, in different illustrative cases. (A) At the rearranged 3L chromosome arm of tel-m1-1. Methylation frequency was plotted over different regions of subtelomere 3L in tel-m1-1 and in CC-4533 as depicted. For the methylation frequency at Chromosome 7 in CC-4533 and tel-m1-1, see also Supplemental Figure S8A. (B) At the broken 14L subtelomere of tel-m2-1, where the internal genomic sequence was then capped by a telomere sequence, compared with CC-4533. (C) At the fused 4L and 4R Sultan arrays of tel-m1-1, compared with the two arrays in CC-4533. (D) At the head-to-head fused 9R Sultan arrays and the transition to rDNA in tel-m1-1, compared with the 9R extremity of CC-4533.











