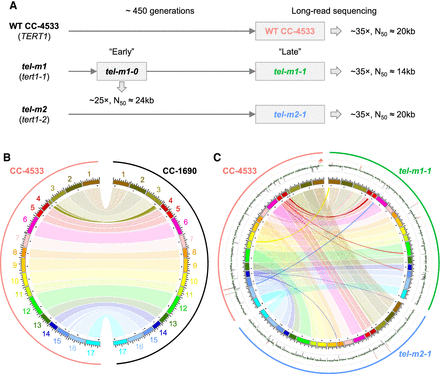
Independent genome assemblies of CC-4533 and telomerase-mutant strains tel-m1-1 and tel-m2-1 reveal few rearrangements at the genome assembly level. (A) Experimental setup and main sequencing output. (B,C) Circos plot (Krzywinski et al. 2009) of chromosome scaffolds from the long-term culture of CC-4533 compared with CC-1690 (B) and with tel-m1-1 and tel-m2-1 (C), shown as colored boxes, with black marks for centromere clusters (composed of the Zepp-L1 repeat). Links connect homologous blocks, colored according to CC-4533 chromosomes. Outer plot displays sequencing depth, averaged over 100-kb regions, with 0×, 35×, and 70× grid lines shown in black, green, and red, respectively. Depth over 50× is highlighted in red, and genomic position is indicated in megabases.











