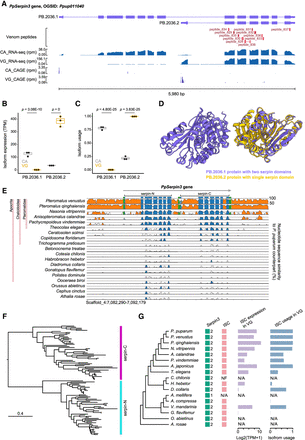
Isoform switching in the serpin3 gene contributes to venom recruitment in P. puparum. (A) Two distinct isoforms (PB.2036.1 and PB.2036.2) of PpSerpin3 are expressed in the venom gland (VG) and carcass (CA). From top to bottom: isoforms, venom peptides supported by mass spectrometry-based proteomics, RNA-seq, and CAGE-Seq data. (rpm) reads per million. For supporting evidence of the venom PB.2036.2 protein, please refer to Supplemental Figure S24. (B) Expression levels of the two PpSerpin3 isoforms in VG and CA, displaying adjusted P-values from the differential expression analysis conducted with DESeq2. (C) Isoform usage of the two PpSerpin3 isoforms in VG and CA, showing the FDR (Benjamini–Hochberg) adjusted P-values from the differential isoform usage analysis. Isoform usage is calculated as expression of a specific isoform divided by the expression of the parent gene. (D) AlphaFold2-generated protein structures of the two PpSerpin3 isoforms. The linker between the two individual domains is not depicted because of challenges in deducing its structure. (E) VISTA sequence conservation plot of the serpin3 gene in Hymenoptera, utilizing P. puparum as the reference. The genomic sequences displaying over 50% sequence identity compared to the reference are shown. Noncoding regions are represented in orange, exons in blue, and UTRs in green. Conserved regions with at least 70% identity over a 100 bp window are indicated in the corresponding color. The gene region, amino-terminal serpin domain (serpin-N), and carboxy-terminal serpin domain (serpin-C) are marked. (F) Maximum-likelihood protein phylogeny of the serpin domain sequences from 51 representative hymenopteran species. (G) Schematic representation of serpin3 genes and their isoforms containing only serpin-C (ISC), accompanied by a species tree of selected representative hymenopteran species. Filled boxes in the “serpin3” column represent genes with intact ORFs, and the numbers beside the boxes indicate the number of serpin domains in the genes. Filled boxes in the “ISC” column indicate the presence of ISC. (NF) not found, (N/A) not applicable. The expression (TPM) and isoform usage of ISCs in VG are visualized in the corresponding bar plots on the right. The N/A for O. abietinus and A. rosae indicates the absence of venom gland RNA-seq data for these two wasps.











