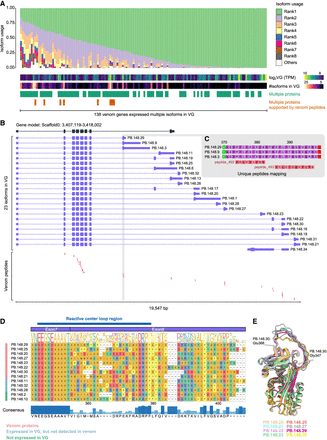
Diverse isoforms of venom genes in P. puparum. (A) Isoform usage of venom genes. Only isoforms expressed in the venom gland are included, and isoforms with TPM values higher than 1 in at least one replicate of the venom gland sample are considered expressed. The usages of isoforms within each gene are ranked (Rank1 represents the most used isoform in a gene) and depicted in different colors. Venom genes with multiple isoforms encoding different proteins are indicated, with emphasis on genes supported by mass spectrometry-based proteomics. This figure showcases venom genes that express multiple isoforms in the venom gland. Please refer to Supplemental Figure S5 for the isoform usage landscape of all venom genes. (B) PpSerpin1 exhibits the expression of 23 isoforms in the venom gland, encoding a total of 12 proteins, with eight of them supported by venom proteome evidence. (C) A zoomed view of the alignments between a protein encoded by isoforms (PB.148.3/.9/.29) and venom peptides. Additional supporting evidence for seven other venom serpin1 proteins can be found in Supplemental Figures S16 and S17. (D) Local alignment of amino acid sequences encoded by PpSerpin1 isoforms starting with Valine343. The putative reactive center loop region is indicated. Isoform PB.148.34 is not included in the alignment because of the absence of the serpin domain. (E) Comparison of 3D protein models predicted using AlphaFold2, highlighting variations in the reactive center loop among the eight venom serpin protein isoforms produced by PpSerpin1 through alternative splicing. The highlighted residues in the PB.148.30 protein structure represent the predicted start and end of the reactive center loop.











