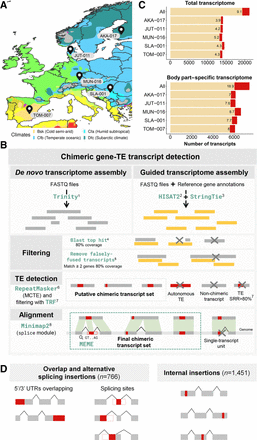
Detection of chimeric gene–TE transcripts in five strains of D. melanogaster. (A) Map showing the sampling locations of the five European strains of D. melanogaster used in this study: TOM-007: Tomelloso, Spain (BSk); MUN-016: Munich, Germany (Cfb); JUT-011: Jutland, Denmark (Cfb); SLA-001: Slankamen, Serbia (Cfa); and AKA-017: Akaa, Finland (Dfc). Colors represent the climate zones according to the Köppen–Geiger climate distribution (Peel et al. 2007). (B) Pipeline to detect chimeric transcripts. (C) Contribution of chimeric gene–TE transcripts to the total transcriptome and the body part–specific transcriptome globally and by strain. (All) All the transcripts assembled in the three body parts and the five strains. (D) Schematic of the two groups of chimeric transcripts identified: overlap and alternative splicing insertion group and internal insertion group. Note that these numbers total more than 1931 because some chimeric transcripts can have different insertions in different genomes. Gray boxes represent exons; red boxes, a TE fragment incorporated in the mRNA; and white boxes, a TE fragment that is not incorporated in the final mRNA. The black lines connecting the exons represent the splicing events.











