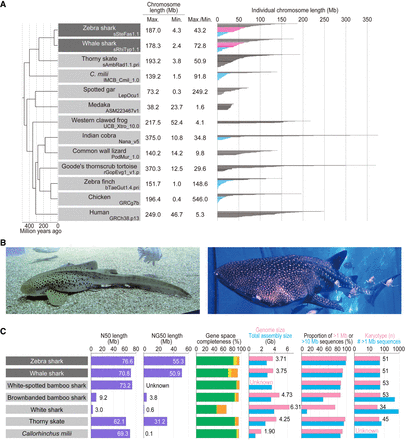
Shark species studied and comparative statistics of their genome assemblies. (A) The karyotypes of diverse vertebrate species are depicted with the length of individual chromosomal DNA sequences. The maximum and minimum chromosome lengths are based on the records in NCBI Genomes. Microchromosomes for osteichthyans are shown in light blue according to individual original reports (International Chicken Genome Sequencing Consortium 2004; Knief and Forstmeier 2016; Suryamohan et al. 2020; Nakatani et al. 2021), whereas the eMID and eMIC of the two shark species whose genomes have been sequenced in the present study (see Results) are shown in magenta and light blue, respectively. (B) Zebra shark (left) and whale shark (right). Photo credit: Shigehiro Kuraku (left), Rui Matsumoto (right). (C) Statistics of the genome assemblies. The identifiers of the genome assemblies, as well as statistical comparison of more metrics, are included in Supplemental Table 1. Gene space completeness shows the proportions of selected one-to-one protein-coding orthologs with “complete” (green), “duplicated” (yellow), and “fragmented” (orange) coverages retrieved in the sequences by the BUSCO pipeline (see Methods). The details of the genome sizes and karyotypes included are based on existing literature (Schwartz and Maddock 1986, 2002; Hardie and Hebert 2004; Uno et al. 2020).











