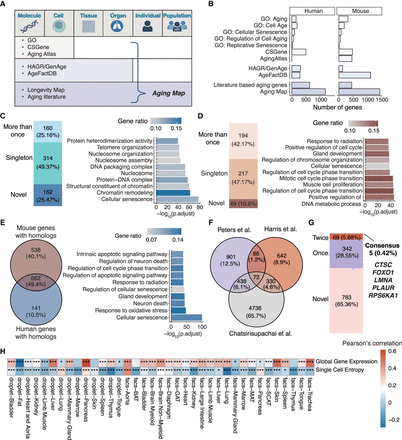
Aging Map: a manually curated database of aging-related transcriptomic features. (A) Summary of data sources used to build Aging Map, grouped by the scales of aging information covered by each collection. (B) Number of aging-related genes collected from each data source. (C) Characteristics of the 636 literature-confirmed aging-related genes in human. (Left) Overlaps with other databases listed in A. (Novel) Not covered in any of the existing database, (singleton) covered by one database, (≥2) covered by at least two databases. (Right) GO Enrichment analysis showing the top 10 GO terms associated with the literature-based aging genes, colored by the proportion of GO gene sets present in the list. (D) Similar to C but showing the results for 460 literature-confirmed aging-related genes in mouse. (E) Comparisons of human and mouse Aging Map genes. (Left) Venn diagram of homologous human and mouse genes. (Right) GO enrichment analysis showing the top 10 GO terms associated with aging genes shared between human and mouse, colored by the proportion of GO gene sets present in the list. (F) Venn diagram comparing human age-associated genes identified in three large cohort studies. For a full description of each data set, see Methods. (G) Overlaps between human Aging Map genes and the three data-driven gene lists in F. The five genes shared by all sets: CTSC, FOXO1, LMNA, PLAUR, and RPS6KA1. (H) Heatmap of correlation between chronological age and two high-level transcriptomic features (global gene expression and single-cell entropy) in each data set. (***) Adjusted P-value < 0.001, (**) adjusted P-value < 0.01, and (*) adjusted P-value < 0.05 using linear regression for global gene expression and Kruskal–Wallis test for single-cell entropy. All P-values were corrected by the Benjamini–Hochberg procedure.











