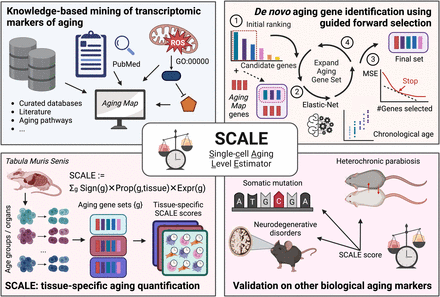
Schematic illustration of SCALE and the study design. We designed SCALE to select interpretable aging-related genes and quantify tissue-specific transcriptomic aging in a robust and easily generalizable manner. SCALE starts with a core set of manually curated aging-associated genes (Aging Map; top left), iteratively expands the list to include additional features that improve age prediction (top right), and eventually uses the gene set to quantify aging for each cell through weighted average of gene expression (bottom left). By explicitly introducing Aging Map genes as seeds to guide feature selection, SCALE encourages the inclusion of both well-documented genes and genes that contain complementary information on aging. SCALE also benefits from its indirect modeling of chronological age, in which the latter is only used to select genes rather than as the final prediction target. This makes SCALE less susceptible to technical noise and batch effects prevalent in single-cell RNA-seq data, allowing it to better capture biological age and the nuanced aging and antiaging effects of disease and interventions (bottom right).











