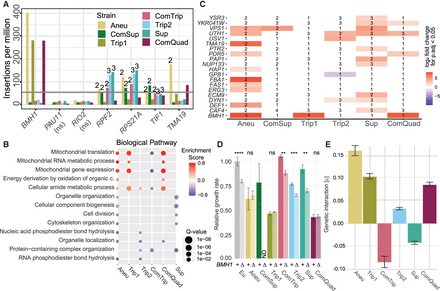
CNV strains have common and allele-specific genetic interactions. (A) Seven genes have no insertions in either replicate of the euploid strain, whereas insertions are identified in these genes in all CNV strains. The gray line represents the minimum insertion count per gene to be considered significant. The numbers above each bar of each strain indicate the copy number of the gene if it is contained within a CNV. If no number is given, it is a single-copy gene. (B) Shared enriched GO terms identified using gene set enrichment analysis (GSEA) of log2 fold-changes obtained from differential analysis comparing each CNV insertion profile to the euploid insertion profiles (Q-value, circle size). For all enriched gene sets, see Supplemental Figure S14. Positive enrichment scores (red) indicate functions that have increased insertion frequencies in the CNV strain, whereas negative enrichment scores (blue) indicate the inverse. ComSup had no significant enrichment of any gene sets. (C) Significant genes (P.adjust < 0.05) from differential analysis comparing each CNV insertion profile to the euploid insertion profiles. Positive values (red) indicate an increase in frequency in the CNV strain; negative values (blue) indicate the inverse. If a gene is amplified, the copy number is annotated. (D) Mean and standard deviation (error bars) of growth rate relative to the ancestral, euploid strain in YPGal batch culture. P-values from two-sample t-test are indicated by the following: (ns) not significant, (*) P < 0.05, (**) P < 0.01, (***) P < 0.001, (****) P < 0.0001. (E) The mean strength and standard deviation (error bars) of the genetic interaction (ε) were determined based on the deviation of expected fitness based on a multiplicative fitness model (Methods) for GAP1 CNV and BMH1 double mutants, calculated from growth rates shown in D.











