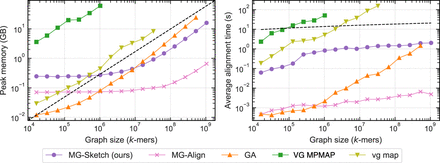
Peak RAM usage and query time versus graph size. Graphs are generated with k = 80 according to the section “Synthetic data generation.” We run MG-Sketch with t = 6, D = 14, w = 16, s = 8, and K = 10 neighbors, using query sequences with a mutation rate of 25%. Traces for VG MPMAP and vg map are incomplete as they exceed time or memory limit. (Left) Peak RAM usage versus graph size, with the black dashed line indicating linear memory complexity. (Right) Average alignment time versus graph size, with the black dashed line indicating logarithmic time complexity. For recall comparisons, see Figure 4.











