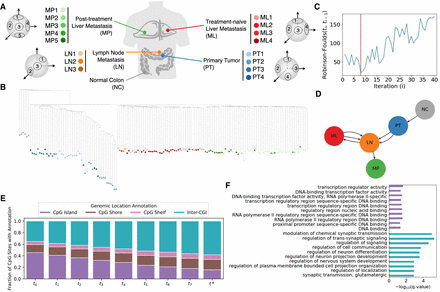
Application of Sgootr to patient CRC01 scTrio-seq data by Bian et al. (2018). (A) The multiregional patient data consist of single cells sampled from four distinct lesions (in addition to normal tissue) and multiple sampling locations within each lesion. (B) Tumor lineage tree constructed by Sgootr with κ=0.1. Each single cell is represented as a leaf, colored by its sampling location. (C) RF distances between the trees of consecutive iterations of the pruning procedure, with the global minimum occurring at t* = t8. (D) Tumor migration history inferred by Sgootr for patient CRC01. (E) Fraction of CpG sites located in CpG island (CGI; regions with overrepresentation of CpG sites) (Cross and Bird 1995), CpG shore (2-kb-long regions flanking both sides of CGIs) (Irizarry et al. 2009), CpG shelf (2-kb-long regions adjacent to CpG shores on the side away from CGIs) (Irizarry et al. 2009), and inter-CGI regions in the CRC01 trees at each iteration of the pruning procedure of Sgootr, from t0 to t*. (F) Top 10 GO terms with significant (<0.05) q-values in enrichment analysis of nonpseudo, protein-coding genes spanning lineage-informative sites in intra-CGI and inter-CGI regions, respectively, in CRC01. For a full list of enriched terms, see Supplemental Figure S12A.











