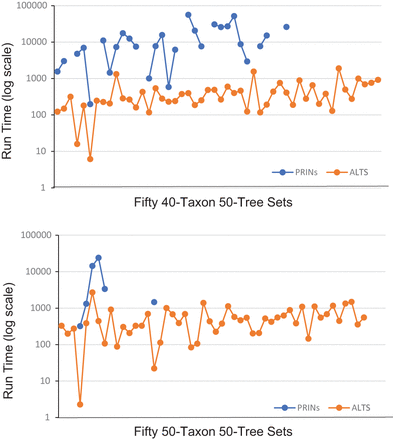
Figure 4.
The run time (in seconds) of ALTS and PRINs on 100 data sets. Each data set contains 50 trees on 40 or 50 taxa. The data sets are sorted in the increasing order according to the HN of the tree–child networks inferred by ALTS. PRINs had some missing data points because of an abort after 24 h of clock time.











