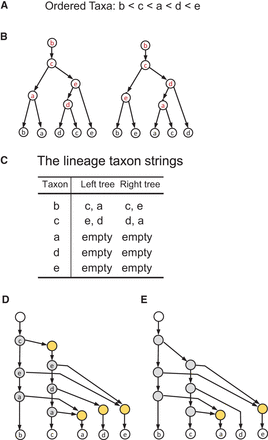
The construction of a tree–child network that displays two phylogenetic trees. (A) An ordering on {a, b, c, d, e}. (B) Two trees, where the internal nodes are labeled w.r.t. the ordering using the Labeling algorithm. (C) The lineage taxon strings (LTSs) of the taxa obtained from the labeling in B. (D) The rooted directed graph constructed from the shortest common supersequences (SCS) of the LTSs of the taxa (in C) using Tree–child Network Reconstruction. The SCS is [c, e, a] for [c, a] and [c, e] and is [e, d, a] for [e, d] and [d, a]. (E) The tree–child network obtained after the removal of the degree-2 nodes.











