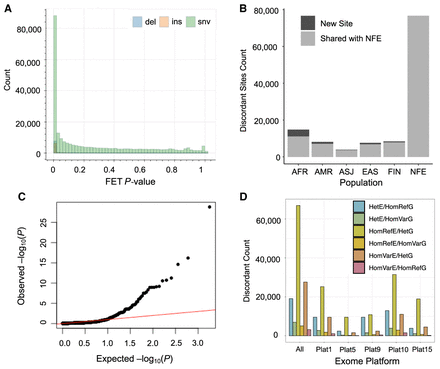
Discordance in genotype calls across high-throughput genotype discovery approaches. (A) Fisher's exact test concordance test P-value for shared, PASS sites in the gnomAD NFE exomes and genomes. Bars are colored by variant type: insertion (ins), deletion (del), or single-nucleotide variant (snv). (B) “Bad” sites are replicated across ancestry groups in gnomAD. Sites flagged as discordant in both the NFE and another ancestry group are plotted in gray; those new sites not in the NFE are shown in black. (C) QQ plot for the Fisher's exact test P-value of shared variants in a set of 946 individuals for whom both WES and WGS data were available. (D) Different exome captures’ contribution to discordant sites. Bars are colored by the error mode that was observed for the discordant genotype call: heterozygous (Het), homozygous reference (HomRef), or homozygous variant (HomVar) in either the exomes (E) or genomes (G).











