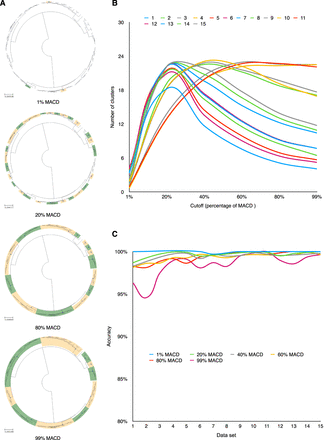
The performance of iterative-PopPUNK at multiple levels. (A) The comparison of iterative-PopPUNK clusters with the genealogical tree (from 100 samples) at four levels: 1%, 20%, 80%, and 99% of the MACD. These green and orange faces represent matched clusters with tree nodes, respectively. For these unmatched clusters, the tree leaves are annotated using different colors. (B) The number of clusters iterative-PopPUNK tree for different cutoff values, averaged over data sets. (C) The accuracy of estimated clusters at each level. The accuracy is calculated by the total number of matched clusters (with tree nodes) divided by the total number of estimated clusters. The chosen example data set has a recombination rate r = 0.006 and mutation rate = 0.01.











