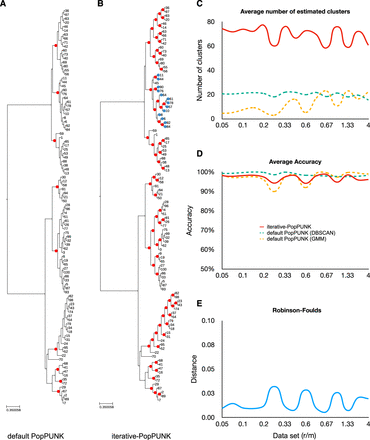
The performance of iterative-PopPUNK on simulated data. The chosen simulated data set for A and B has a recombination rate r = 0.001 and mutation rate μ = 0.01. (A) A genealogical tree with nine nodes (clusters) estimated by default PopPUNK. (B) The same genealogical tree with 69 clusters estimated by iterative-PopPUNK, among which 68 can be totally matched with genealogical tree nodes although one of them cannot be matched, with strains in the unmatched cluster shown in blue. (C) A line graph shows the average number of estimated clusters by PopPUNK and by iterative-PopPUNK. The solid lines present the results for iterative-PopPUNK, and the dash lines are for default PopPUNK. (D) The accuracy results for both default PopPUNK and iterative-PopPUNK. The accuracy is calculated from the total number of matched nodes divided by the total number of estimated clusters. (E) Robinson-Foulds distances between true trees and iterative-PopPUNK trees. r/m stands for recombination rate divided by mutation rate.











