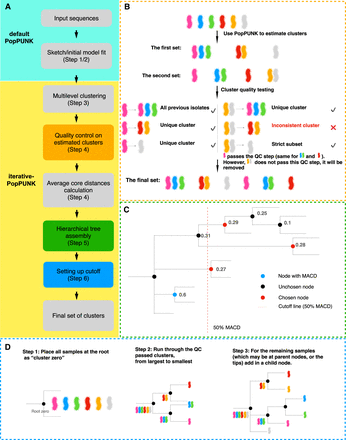
Workflow for developing iterative-PopPUNK. (A) Steps for designing iterative-PopPUNK. After inputting sequence data, PopPUNK creates a local sketching database, which is further used to estimate clusters. QC steps for removing inconsistent clusters are shown in the orange box. The green box shows how to nest these estimated clusters into an iterative-PopPUNK tree. Methods for choosing the final set of clusters are presented in the blue box. (B) QC algorithm. Three conditions for determining QC-passed clusters are described in detail in Methods, which in short are as follows: (1) The new clusters contain all of the isolates from previous clusters; (2) the new cluster is unique and contains none of the items from previous clusters; (3) the new cluster is a strict subset of one of the previous clusters. (C) Algorithm for cutting iterative-PopPUNK tree. The goal is to choose the closest node to the cutoff line but with a smaller value. In this example, the cluster (node) annotated using blue color has the maximum value of average core distances (0.6). The red dashed line shows the cutoff is 50% of the MACD (0.3). Therefore, the node with an ACD lower than and closest to 0.3 will be selected. (D) Hierarchical tree assembly. The dashed lines indicated these potential branches during the tree assembly process.











