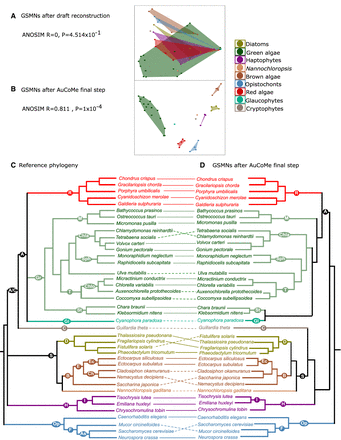
AuCoMe as a tool to improve taxonomic consistency of GSMNs. (A,B) MDS plots for GSMNs calculated with the AuCoMe draft reconstruction step (A) or after all AuCoMe steps (B). In both cases, ANOSIM values are indicated below (MDS and ANOSIM were computed using the vegan package [https://github.com/vegandevs/vegan] with R 4.1.2 [R Core Team 2023]). (C,D) Tanglegram evaluating the taxonomic consistency between reference phylogeny, compiled from Strassert et al. (2021) (C) with AuCoMe dendrograms based on metabolic distances using the pvclust package version 2.0.0 (Suzuki and Shimodaira 2006) with R 3.4.4 (R Core Team 2023) with the Jaccard distance (D). Full lines join species for which the position in the AuCoMe dendrogram is consistent with the reference phylogeny. Dotted lines join species for which the metabolic dendrogram and the reference phylogeny diverge. (A/C) Archeplastids/cryptophytes, (A) archeplastids, (R) rodophytes, (Gr) green algae, (M) Mamiellales, (Chla) Chlamydomonadales, (Sph) Sphaeropleales, (T) Trebouxiophyceae, (Chlo) Chlorellaceae, (St) streptophytes, (Gl) glaucophytes, (C) cryptophytes, (H) haptophytes, (I) Isochrysida, (D) diatoms, (S) Stramenopiles, (B) brown algae, (E) Ectocarpales, (Ec) Ectocarpaceae, (Ch) Chordariaceae, (Op) opistochonts, (F) fungi, (As) ascomycetes.











