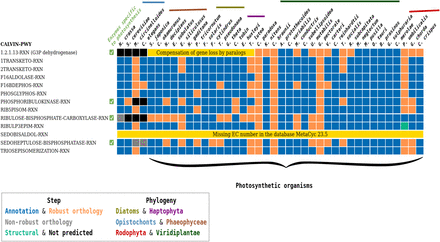
AuCoMe results on the Calvin cycle pathway in the algal data set. AuCoMe was applied to the data set of 36 algae and four outgroup species (columns). Each row represents a MetaCyc reaction of the pathway; the table shows whether it is predicted by AuCoMe: blue, draft reconstruction; orange, robust reactions predicted by orthology propagation that passed the filter; green, structural verification; gray, nonrobust reactions predicted by orthology propagation and removed by the filter; black, not predicted; and yellow, manually added because the MetaCyc database 23.5 does not contain a reference gene–reaction association for this reaction.











