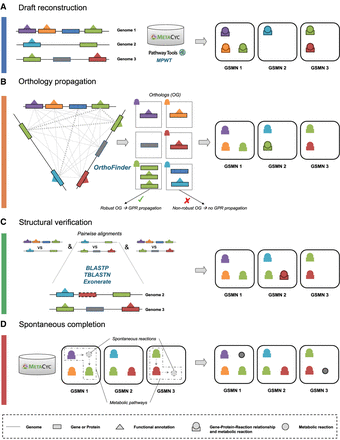
Reconstruction and homogenization of metabolisms with AuCoMe. Starting from a data set of partially structurally and functionally annotated genomes, AuCoMe's pipeline performs the following four steps. (A) Draft reconstruction. The reconstruction of draft genome-scale metabolic networks (GSMNs) is performed using Pathway Tools in a parallel implementation. (B) Orthology propagation. OrthoFinder predicts orthologs by aligning protein sequences of all genomes. The robustness of orthology relationships is evaluated (see Methods), and gene–protein–reaction (GPR) of robust orthologs are propagated. (C) Structural verification. The absence of a GPR in genomes is verified through pairwise alignments of the GPR-associated sequence to all genomes where it is missing. If the GPR-associated sequence is identified in other genomes, the gene is annotated, and the GPR is propagated. (D) Spontaneous completion. Missing spontaneous reactions enabling the completion of metabolic pathways are added to the GSMNs. (OG) Orthologs. Outlines around GPR or reaction indicate that the GPR or reaction is newly added during the corresponding step.











