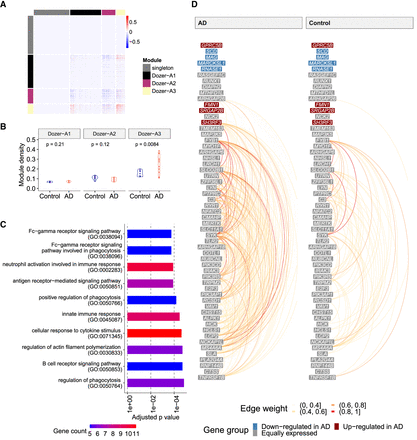
Coexpression network analysis of the Morabito_2021 multiple donor scRNA-seq data. (A) Heatmap of the “difference network” (average of AD networks − average of control networks). Genes are ordered by gene modules (singleton, Dozer-A1, Dozer-A2, Dozer-A3), which are obtained by hierarchical clustering on the “difference network” (Methods). (B) Violin plots of module densities for AD and control donor networks. Module density is a function of the average absolute correlation of gene pairs in the given module (Methods). (C) Bar plot of GO enrichment terms for module Dozer-A3. (D) Hive plot visualization of module Dozer-A3 in AD and control groups. Genes are ordered from high (top) to low (bottom) by their average expression across all donors. Genes are further divided into three groups as “down-regulated in AD,” “up-regulated in AD,” and “equally expressed” according to their differential expression status between the control and AD diagnoses. The arcs between the genes in this linear layout depict the edges in the average networks of AD (left) and control (right) donors, with colors representing edge weights.











