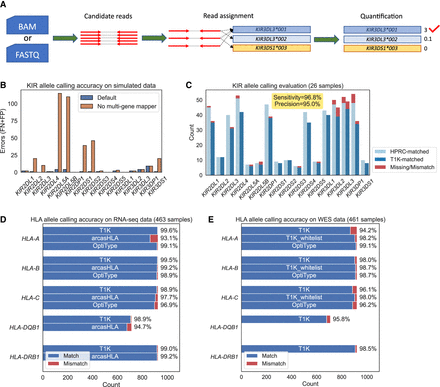
Evaluation of T1K. (A) Overview of the T1K workflow. (B) The KIR allele prediction accuracy of T1K and T1K with no multiple-gene mapped reads (mimicking arcasHLA) on simulated reads from KIR mRNAs. (FN) False negative; (FP) false positive. (C) The KIR allele prediction accuracy of T1K on WGS. (HPRC matched) The alleles inferred from the HPRC phased genome that can be found in T1K predictions; (T1K matched) the alleles predicted by T1K that can be found on HPRC genomes. (D) The HLA allele prediction accuracy of T1K, arcasHLA, and OptiType on RNA-seq data validated with 1kPG annotation. (E) The HLA allele prediction accuracy of T1K and OptiType on WES data validated with 1kPG annotation. With T1K_whitelist, T1K only reports alleles present in an allele whitelist coded in OptiType's Python script.











