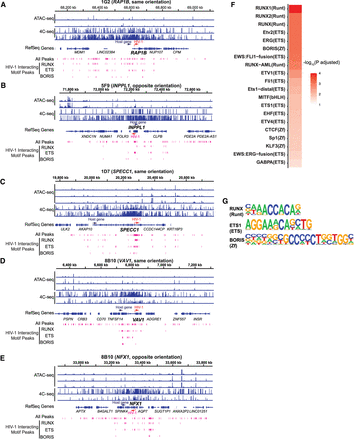
Transcription factor binding motifs enriched in both increased chromatin accessibility and HIV-1–host chromatin interactions. (A–E) HIV-1 interacting peaks, defined as regions of overlapping ATAC-seq peaks and 4C-seq signal (fragments detected were 10 or more in at least one replicate out of 40 possible), were extracted from the integrated chromosome in each cell line. (F) Transcription factor binding motifs that were enriched in HIV-1 interacting peaks were identified by HOMER. Top 20 known motifs ranked by adjusted P-values. (G) Example motif logos for each of the top three transcription factor groups: ETS, RUNX, and ZNF. P-values were corrected for multiple hypothesis testing by the Benjamini–Hochberg procedure.











