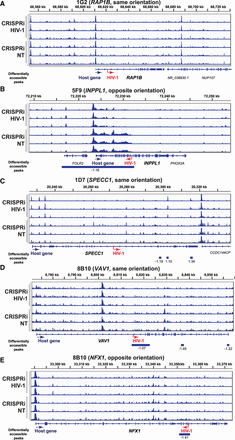
HIV-1 promoter activity repression reduces host chromatin accessibility near the HIV-1 integration site. (A–E) CRISPRi-mediated HIV-1 promoter inhibition reduces host chromatin accessibility near the integration site. ATAC-seq tracks were normalized by counts per million (CPM) scaled by the fraction of reads in peaks and display a data range of zero to three. Each peak is annotated with its log2 fold change relative to the nontargeting control. HIV-1 sgRNA targets HXB2 coordinates 344–363 in HIV-1 LTR promoter. (NT) Nontargeting gRNA. Statistics performed by DESeq2; all peaks displayed have an adjusted P-value < 0.05.











