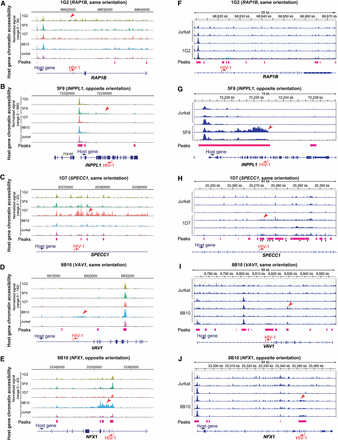
HIV-1 increases host chromatin accessibility ∼5–30 kb around the HIV-1 integration site. (A–E) Host chromatin accessibility captured by single-cell DOGMA-seq. (F–J) Host chromatin accessibility captured by bulk ATAC-seq. (A–E) Normalized by Signac. (F–J) Normalized by counts per million (CPM) and displaying a data range of zero to one. An asterisk denotes adjusted P-values < 0.05 by DESeq2 when comparing uninfected Jurkat T cells to the indicated HIV-1-infected clonal cell line. Red arrowheads indicate examples of increased host chromatin accessibility identified in respective cell line clones. Peaks shown were called by MACS2 for DOGMA-seq cells and by Genrich for bulk ATAC-seq.











