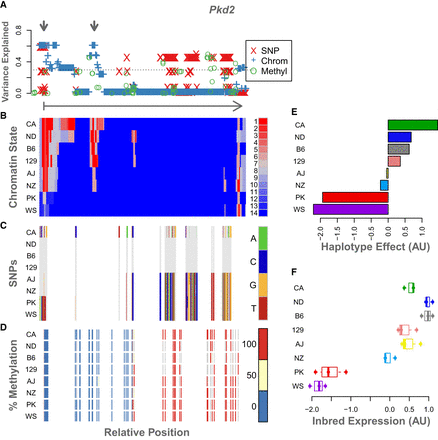
Example of epigenetic states and imputation results for a single gene, Pkd2. The legend for each panel is displayed to its right. (A) The variance in DO gene expression explained at each position along the gene body by each of the imputed genomic features: SNPs, red X's; chromatin state, blue plus signs; and percentage of methylation, green circles. The horizontal dotted line shows the maximum variance explained by any individual haplotype (in this case, CAST). For reference, the arrow below this panel runs from the TSS of Pkd2 (vertical bar) to the TES (arrowhead) and shows the direction of transcription. The gray arrows at the top indicate two regions of interest where chromatin state explains height amounts of variance in gene expression. (B) The chromatin states assigned to each 200-bp window in this gene for each inbred mouse strain. States are colored by their association with gene expression in the inbred mice. Red indicates a positive association with gene expression, and blue indicates a negative association. Each row shows the chromatin states for a single inbred strain, which is indicated by the label on the left. (C) SNPs along the gene body for each inbred strain. The reference genotype is shown in gray. SNPs are colored by genotype as shown in the legend. (D) Percentage of DNA methylation for each inbred strain along the Pkd2 gene body. Percentages are binned into 0% (blue), 50% (yellow), and 100% (red). (E) Association of haplotype with expression of Pkd2 in the DO. Haplotype effects are colored by the allele from which they were derived. (F) Pkd2 expression levels across inbred mouse strains. For ease of comparison, panels B through F are shown in the same order as the haplotype effects.











