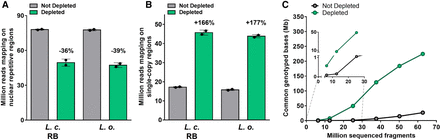
Performances of CRISPR-Cas9-mediated repeat depletion in different lentil samples. (A,B) Sequencing reads mapping to the repetitive or single-copy regions with or without CRISPR-Cas9-mediated repeat depletion in multiplex libraries generated from different lentil samples, namely L. culinaris cv. Redberry (L. c. RB) or L. orientalis (L. o.). Data are means ± SE (n = 2) normalized for the same sequencing input (50 million fragments). Variation percentages observed following CRISPR-Cas9-mediated repeat depletion are reported above each condition. (C) Number of genotyped positions in common between all samples analyzed in the study (three distinct samples of L. culinaris cv. Castelluccio, one sample of L. culinaris cv. Redberry, and one sample of L. orientalis), within the single-copy regions. N = 5 at 6, 12, and 25 million fragments; N = 4 at 37, 50, and 63 million fragments.











