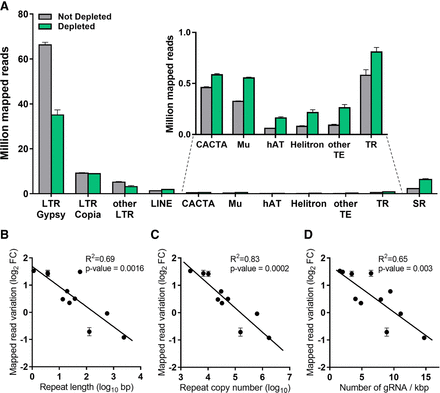
Sequencing data distribution after CRISPR-Cas9-mediated repeat depletion according to the class of nuclear repeat. (A) Number of reads (in millions) mapping to different nuclear repeat classes with or without CRISPR-Cas9-mediated repeat depletion: Ty3-Gypsy (LTR Gypsy), Ty1-Copia (LTR Copia), other LTR, LINE, CACTA, Mu, hAT, Helitron transposons, other transposable elements with abundance < 0.1% (Harbinger, mariner, Sine), tandem repeats (TR), and simple repeats (SR). Correlation between the variation of mapped reads following CRISPR-Cas9-mediated repeat depletion on the same repeat classes versus the repeat length (B), repeat copy number (C), and density of gRNAs targeting each repeat class (D). Data are means ± SE (n = 3). (FC) Fold change.











