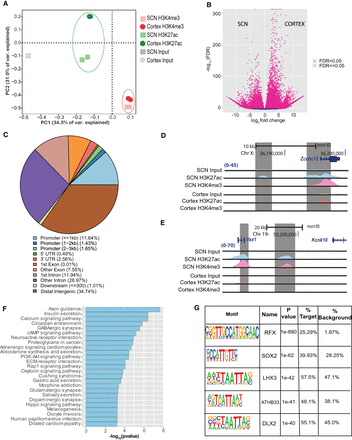
Genome-wide characterization of SCN enhancers. (A) PCA analysis of aligned reads post H3K4me3 and H3K27ac immunoprecipitation from the SCN and cortex mouse brain tissues at ZT3. (B) Volcano plot showing fold change and false-discovery rate (FDR) for differential H3K27ac sites between the SCN and cortex as computed by Diffbind. (C) Genomic feature distribution of SCN-enriched H3K27ac peaks (n = 14,153). (D,E) UCSC Genome Browser tracks showing histone modifications H3K4me3 and H3K27ac normalized ChIP-seq read coverage (shaded gray) along with their input for the SCN and cortex at representative examples. The chromosome location and scale (mm10 genome) are indicated at the top. (F) Functional annotation of nearest neighboring gene (TSS) to SCN-enriched H3K27ac sites (fold change > 5) using the KEGG pathway (DAVID). (G) Overrepresented transcription factor binding motifs using SCN-enriched H3K27ac sites as target by HOMER.











