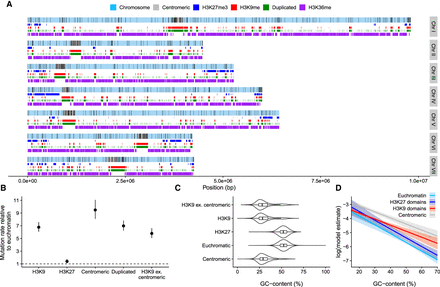
Variation in mutation rate across the genome. (A) Distribution of mutations along the seven chromosomes; black lines indicate mutations. Centromeric regions, H3K27 trimethylation, H3K36 dimethylation, H3K9 trimethylation domains, and duplicated regions are shown. (B) Relative mutation rates for different genomic domains. H3K9 ex. centromeric are H3K9me3 domains in which overlaps with centromeric regions have been excluded. Posterior medians and 95% HPD intervals are shown. (C) Violin plots overlaid with boxplots for GC-content in different domains. (D) Model estimates (on a log-scale) for the mutation rate from a model with GC-content, H3K9me3, H3K27me3, and centromeric domains as predictors. Lines are posterior medians, and envelopes are 95% HPD intervals.











